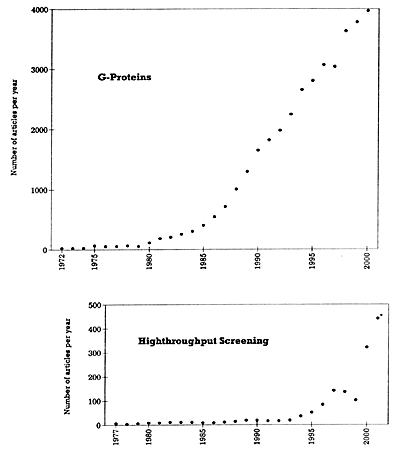
* The 2001 entries are based on the number of articles published in the first 5 months and then projected for the whole year.
| Preskorn.com | Printed from: http://www.preskorn.com/columns/0109.html |
| Drug Development in Psychiatry and the Human Genome Project: The Explosion in Knowledge and Potential Targets |
|
|
|
SHELDON H. PRESKORN, MD |
|
Journal of Psychiatric Practice, September 2001, 336-340 |
The goal of this series of columns is to explain 1) the process of drug development and its relevance to the optimal practice of clinical psychopharmacology and psychiatry, and 2) the implications of the human genome project for future drug development in psychiatry. The last column discussed the process of "drilling down" on the target of interest.1 This column will extend that discussion by reviewing the explosion in knowledge relevant to drug development in general, and psychiatry in particular, Results from PubMed searches will be presented illustrating the rapid expansion of biological knowledge and chemical techniques relevant to modern drug development that has occurred over the last 25 years.
Recall that the following equation is central to all clinical pharmacology:
Effect = affinity for x drug x biological
site of action concentration variance
(Equation 1)
The first variable in this equation, the site of action, determines the potential effects that the drug can produce. By binding to that site of action, the drug can alter physiological mechanisms regulated by that site of action. Parenthetically, the affinity of the drug for the site of action determines the concentration (i.e., the second variable in the equation) needed to engage (i.e., agonist effect) or disengage (i.e., antagonist effect) the site of action to a physiologically and hence clinically meaningful degree. The third variable is the biological variance among humans that may shift tile dose-response curve for a given individual (i.e., the "sensitive" or "resistant" patient).
Antibiotics as a Model for Drug Discovery
As discussed in previous columns in this series, the first stage of drug development is drug discovery. In the beginning, this process was highly dependent on chance (Stage 1). It began with observations dating back to antiquity about the health enhancing properties of herbs and other natural products. Of relevance to psychiatry, such observations included the "mind altering" properties of naturally occurring substances such as alcohol, LSD, marijuana, opium, peyote, and other plant products. The fact that such substances could alter cognition, perception, and even personality has been noted in the earliest records of human history.
Drug discovery received a significant boost with Fleming's chance discovery of pencillin and the subsequent outbreak of World War IL Like the Manhattan project to develop the atomic bomb, World War II created the demand and the funding to mass produce antibiotics to treat battlefield infections.
The development of antibiotics has served as the general model for most drug development in the 20th century. The driving principle has been to understand how the drug had to affect a target to produce the desired clinical outcome. As previously discussed, the first approach to modern drug development (Stage II, 1900-1940s) was to develop newer drugs by altering the structure of drugs discovered by chance.2 The next approach (Stage 111, 1950s-present) was to isolate and characterize the target of interest and develop compounds that specifically interacted with that target.
Although nothing about drug development is easy, the development of anti-infectives, even for the Human Immunosuppression Virus (HIV), is relatively easier than developing drugs to treat brain diseases. There are multiple reasons for this fact.
Isolating and Studying the Target of Interest
The first reason is that, when studying infectious diseases, one can relatively readily isolate and study the target of interest (i.e., generally a regulatory protein fundamental to the reproduction or viability of the bacteria or the virus) as well as the effect of drugs on the target. One can, after all, grow the bacteria or virus in the laboratory, study it to determine which of its subcomponents are likely targets for drug development, and then study the effect of specific drugs on that target and in turn on the intact bacteria or virus.
| Table 1. Number of publications found in PubMed by key words of relevance to the human genome project and drug development in psychiatry | |||||
| |||||
A fascinating case in point is the development of protease inhibitors to prevent the production of infectious HIV.3-5 What follows is a simplified summary of the strategy used to develop this class of anti-infectives. The approach was to study the various steps in the replication of the virus and determine what were likely steps that could be altered by a drug in such a way as to prevent the production of infectious virus. A protease enzyme was identified that cleaved the provirus into the infectious virus, which could then be encapsulated and disseminated. After identification, the structure of this enzyme was characterized with particular attention to the active site responsible for cleavage of the provirus. Based on this knowledge, new molecular entities (NMEs) could be designed to occupy this site and inhibit its ability to cleave the provirus. That is the essence of establishing structureactivity relationships (SARs).6,7 In this case, the desired activity was inhibition of this enzyme (i.e., the desired target). The desired NME structure was the basic configuration needed to occupy the enzyme and prevent it from being able to appropriately cleave the provirus.
Armed with the knowledge of the needed structure, the next step was to determine whether an NME could be developed that had the necessary structure and also the properties needed to be a drug. Those properties include adequate bioavailability so that it can reach the target of interest, adequate residual time in the body to be effective, and then adequate safety, tolerability, and finally efficacy. Note that the preceding description summarizes all the stages of drug development.7
An Alien Versus a Human Target of Interest
The second reason why anti-infectives are relatively easier to develop than other drugs is that their target is alien (i.e., not from a human), so that the potential exists to develop anti-infective drugs that substantially affect the alien target without cross-reacting with any critical human targets. All other drug classes-including drugs for psychiatric illnesses-are designed to affect human sites of action (i.e., targets) and thus alter human physiology. Most drugs, other than anti-infectives, have a human regulatory protein as their site of action (i.e., the first variable in equation 1). These regulatory proteins include various receptors, enzymes, and/or ion channels. By altering the functional status of a regulatory protein, these drugs modify human physiology.
As the saying goes, therein lies the rub. This is the reason why other classes of drugs have a narrower therapeutic index compared to antibiotics and other anti-infectives. In fact, excessive effects on these human regulatory proteins mediate many of the adverse and even toxic effects of such drugs. These include the extrapyramidal side effects (EPS) of selective dopamine-2 receptor blockers (e.g., haloperidol) and the various serotonin-mediated adverse effects of serotonin reuptake inhibitors (e.g., fluoxetine). The pharmacological principles underlying this last statement have been discussed in previous columns.8-11 Case examples of adverse effects resulting from excessive blockade of these respective targets by some of these drugs have also been the subject of previous columns.12-14
Since regulatory proteins share characteristics, even across species as different as yeast and man, there is the potential for an anti-infective to cross-react with a human regulatory protein. A fascinating example of this is the story of azole antifungals such as ketoconazole. These drugs were developed to inhibit an enzyme that produced a constituent necessary for stable cell walls in yeast.15-17 Again, the scientist in drug discovery can isolate and grow the offending organism in the laboratory to study its biochemistry and physiology. A target critical to the life cycle of the offending organism can be identified and its structure characterized. Based on this knowledge, NMEs can be designed with the goal of finding a new drug. All of that occurred with azole antifungals. Unfortunately, the structure that gave ketoconazole the ability to inhibit the yeast enzyme cross-reacted with and inhibited the human cytochrome P450 (CYP) enzyme 3A3/4.16 That fact set the stage for the potentially fatal interaction between ketoconazole and terfenadine because CYP 3A3/4 is a major drug metabolizing enzyme and plays an important role in the first pass metabolism of drugs such as terfenadine.18 Parenthetically, CYP 3A3/4 is an evolutionarily important enzyme that keeps ingested foreign substances, originally xenobiotics and later drugs, from passing into the body from the bowels.19-21
 * The 2001 entries are based on the number of articles published in the first 5 months and then projected for the whole year. | |
| Figure 1. Number of articles published per year on G-proteins and highthroughput screening |
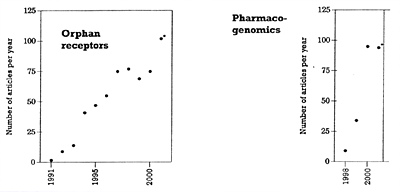 * The 2001 entries are based on the number of articles published in the first 5 months and then projected for the whole year. | |
| Figure 2. Number of articles published per year on orphan receptors and pharmacogenomics |
From Alien to Human Targets
The effects of plant products, such as digoxin and opium, made it clear that NMEs could be developed that can alter human physiology in ways that relieve human suffering and treat disease. The question became: "How can one adapt the process which had been so successful in developing anti-infectives to develop drugs to treat human disease by altering human physiology?" The answer was to identify the targets of interest (i.e., variable 1 in equation 1) and study them in isolated systems before proceeding to the whole human organism.
Oncology, particularly hematology, is an area of drug development where the target, even though of human origin, can be viewed as somewhat analogous to alien microbes. After all, the cancer is a human cell gone awry. It no longer recognizes its boundaries but instead grows out of control. Many of these cells can be studied in isolation in much the same way that microbes can be grown and studied in the laboratory. In a similar way, the drug development scientist can test in vitro whether a NNE has the capability to kill the cancer cell. However, the targets that mediate that toxic effect are human. For that reason, cancer chemotherapy is frequently toxic to a variety of other human cell types. The toxicity is typically most pronounced in cells that turn over rapidly, such as granulocytes. More recently, some success has been achieved by using findings from molecular biology to more specifically target the mechanisms that make cancer cells cancerous, thus improving the drugs' therapeutic index.
While further discussion of drug development in modern cancer chemotherapy is beyond the scope of this column, these developments do illustrate the continuity between psychiatric drug development and drug development in other therapeutic areas ranging from anti-infectives to cancer chemotherapy to drugs targeting physiology within more complex organ systems.
From Single Cells to Complex Systems
The simpler the system being studied, the easier it is to isolate and study it in the laboratory. Nevertheless, it is still by no means as easy as developing anti-infectives for all the reasons discussed above. The gastrointestinal (GI) system, the cardiovascular system (CVS), the kidneys, and the lungs are organs/systems that are neither simple nor easy to study, except when compared to the brain. These organs/systems have fewer molecular components, and the arrangement of these components is much simpler and can be more readily isolated and studied in vitro. For example, isolated guinea pig ileum preparations were used over 50 years ago to study the effects of anticholinergic drugs and other agents on GI motility. The same was done with the CVS, using isolated preparations of heart tissue, arterioles, capillaries, and venules.
Such preparations can be used in drug discovery and also in the more extensive preclinical phase of drug development.7 The drug discovery scientist can search for targets of interests within these in vitro systems and can then use the systems to test and refine the SAR paradigms they are using to guide the synthesis of NMEs (e.g., beta blockers, calcium channel blockers). Once an NME with the desired characteristics has been developed, it can be taken into the preclinical phase of drug development to further determine whether it has the properties necessary to be a commercially viable drug.
While there are in vitro preparations that are used to study the effects of drugs on brain mechanisms, they are much more limited than those used to study other organ systems. These have historically included "grind and bind" receptor assays, synaptosome preparations, and isolated brain slices, to name but a few. However, all of these systems fail to capture the highly integrated nature of brain physiology or its relationship to the output of interest. The latter is a particularly vexing problem for psychiatric drug development, where the outputs of interest include perceptions, motor function, emotions, and cognitions (i.e., the effects in equation 1).
Identifying New Targets Through Molecular Biology
There has been a veritable explosion in the amount of biologically meaningful data and techniques available to the drug discovery scientist. To illustrate, several literature searches were performed using the National Library of Medicine's PubMed site22 to quantitate the number of publications in various areas relevant to modern drug development and to see to what extent publications in these areas have increased over the last 25 years. Parenthetically, the number of publications in an area does not necessarily correlate one-to-one with knowledge. Nevertheless, most readers will likely concede that there is some correlation between the number of peer-reviewed publications in quality journals in specific areas of science and the extent of knowledge in that area.
With the above caveat in mind, Table 1 shows the number of publications that can be found in PubMed using key words that are relevant to the human genome project and potentially to psychiatric drug development. Further searches were done in four of these areas that are particularly relevant to the topic of drug development in psychiatry: G-proteins, highthroughput screening, orphan receptors, and pharmacogenomics.7 The results of these four searches are shown in Figures 1 and 2 to illustrate the dramatic and accelerating explosion in the numbers of publications in these areas. In all four of these specific areas, the accelerating pace has been most pronounced over the last 5 years and will likely increase even more in the future. This explosion in knowledge has significant implications for drug discovery in psychiatry, and for the training of psychiatrists and for their area of expertise relative to other health care workers. It will likely become increasingly difficult for those without special training in biology, biochemistry, and pharmacology to stay abreast of developments in these areas.
Subsequent columns in this series will discuss how knowledge gained through the human genome project and molecular biology in general is increasing the number of potential clinically relevant targets for drug development in psychiatry and how that is affecting the human phases of psychiatric drug development.